Microbial Heterogeneity and Imaging
We investigate how bioprocess conditions foster microbial heterogeneity in bioreactors and develop imaging and other analytical tools for real-time sensing.
Scale-up process development is often seen as more of an art than a science (Humphrey, 2008; Stocks, 2013). Lab-scale tests in microtiter plates are subject to capillary effects and thereby do not represent scale-up performance that involves turbulence and heterogeneities (see figure below). Large scale reactor studies can be resource intensive, minimizing the number of studies that can be performed.
A statistical design-of-experiments approach can help in achieving statistical confidence, but only in a limited portion of the vast multi-parametric experimental design space. This empirical approach leads to low representation of the experimental space.
Minimal replication leads to a restricted understanding of scale-up performance and thereby increases the risk of failure during scale-up of bioprocesses (Crater and Lievense, 2018).
We are establishing scientific methods needed to accelerate fermentation process development in bioreactors.
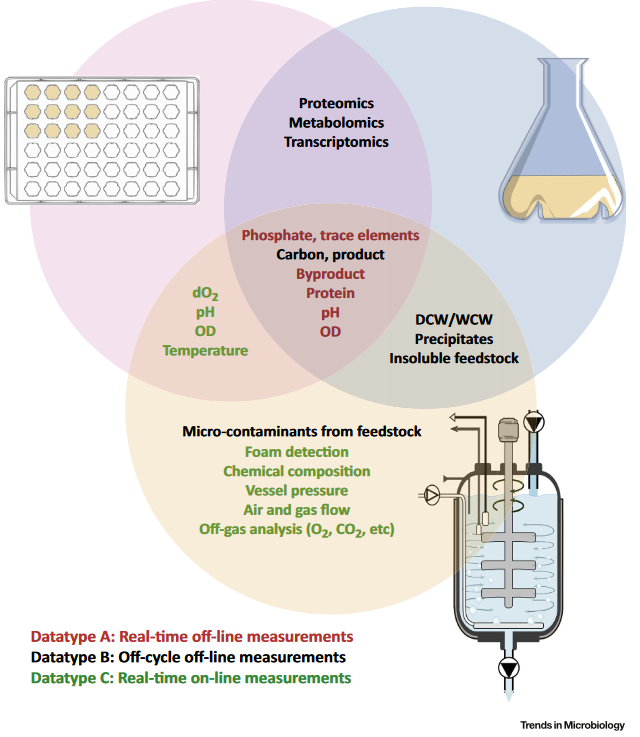
Lab-scale and high-throughput replications have consistently shown much lower variation among replicates, representing only natural stochasticity. In bioreactors, culture conditions could be resonating with natural stochasticity, operational deviations, or both, leading to heterogeneity in culture genetics. System-level heterogeneity starts at the inoculum level and rapidly amplifies by the time of inoculation, leading to high levels of variability. We are ascertaining the evolution of microbial heterogeneity in bioreactors with changing process conditions. We are also developing real-time and off-line methods, including imaging, to rapidly detect the impact of process conditions on intracellular conditions.
Ascertaining the “rules” of microbial heterogeneity with process conditions and measuring them in real-time gives us the opportunity to equip a bioreactor with AI and ML methods and enable adaptive bioprocess optimizing, i.e. in a self-driving mode. Self-driving bioreactors will significantly improve testing efficiency and thereby expand testing to parameter space typically neglected due to resource constraints. This “science of scale” project will reduce scale-up risk and maximize product yields, even at the commercial scale, delivering sustainable aviation fuels and other important bioproducts at competitive prices.
Featured Projects
- “Elucidating Causes of Heterogeneity in Engineered Yarrowia lipolytica.” Collaborators: University of Delaware, Washington University, and Pacific Northwest National Laboratory. Funded by the Department of Energy, BioEnergy Technologies Office through Agile Biofoundry Directed Funding Opportunity 2020.
- “Porting synthetic addiction circuits to reduce evolving culture heterogeneity in non-model Organisms.” Collaborators: Endurogenetics, Inc., National Renewable Energy Laboratory, and Los Alamos National Laboratory. Funded by the Department of Energy, BioEnergy Technologies Office through Agile Biofoundry Directed Funding Opportunity 2020.
- “Quantum Imaging to Measure Metabolite Dynamics in Bioreactor Cultures.” Collaborators: Earth and Environmental Sciences Area, Lawrence Berkeley National Laboratory and Department of Chemistry, UC Berkeley. Funded by Lawrence Berkeley National Laboratory through the Lab Directed Research and Development program. 2020
- “Round-Robin and Reproducibility.” Collaborators: Sandia National Laboratory, National Renewable Energy Laboratory, and Pacific Northwest National Laboratory. Funded by the Department of Energy, BioEnergy Technologies Office through Agile Biofoundry Process Integration and Scale-Up Task. FY18-21.
Featured Publications
- Banerjee, D., Eng, T., Lau, A.K., Sasaki, Y., Wang, B., Chen, Y., Prahl, J-P, Singan, V.R., Herbert, R.A., Liu, Y., Tanjore, D., Petzold, C.J., Keasling, J.D., and Mukhopadhyay, A. 2020. Genome-scale metabolic rewiring improves titers rates and yields of the non-native product indigoidine at scale. Nature Communications 11: 5385
- Wehrs, M., Thompson, M.G., Banerjee, D., Prahl, J-P, Morella, N. M., Barcelos, C. A., Moon, J. A., Costello, Z., Keasling, J. D., Shih, P. M., Tanjore, D.*, and Mukhopadhyay, A. 2020. Investigation of Bar-seq as a method to study population dynamics of Saccharomyces cerevisiae deletion library during bioreactor cultivation. Microbial Cell Factories 19: 167
- Wehrs, M., Tanjore, D., Eng, T., Lievense, J., Pray, T.R., Mukhopadhyay, A. 2019. Engineering Robust Production Microbes for Large-Scale Cultivation. Trends in Microbiology. 27 (6): 524-537
- Wehrs, M., Prahl, J.-P., Moon, J., Li, Y., Tanjore, D., Keasling, J.D., Pray, T., Mukhopadhyay, A. 2018. Production efficiency of the bacterial non-ribosomal peptide indigoidine relies on the respiratory metabolic state in S. cerevisiae. Microbial Cell Factories 17(1): 193