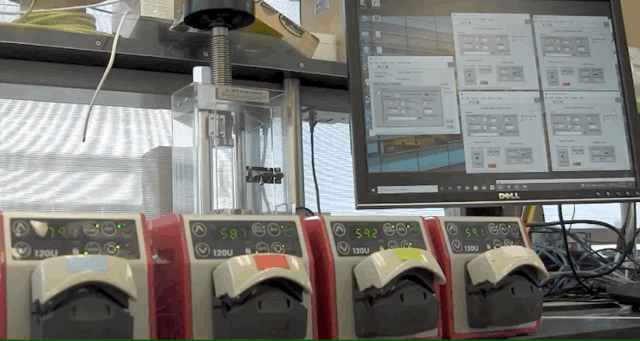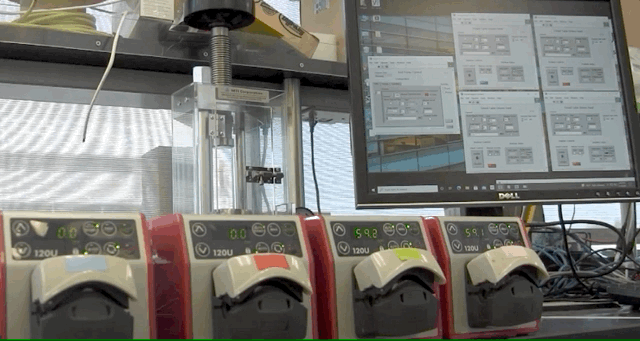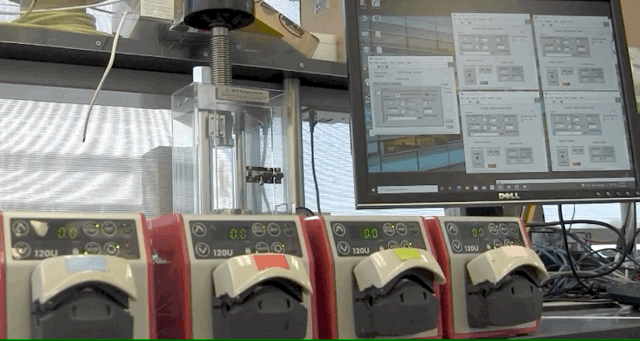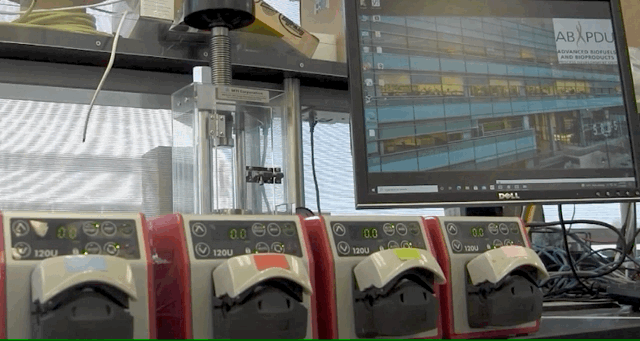
ABPDU has developed an open-source software application that can help universities and early stage biotech companies run fed-batch fermentations, eliminating the need for expertise in controls engineering.
The software solution resulted from a collaboration with Laney College’s Biomanufacturing Program. Douglas Bruce, an instructor in the program, reached out to ABPDU earlier this year in search of a solution that would enable their students to operate bioreactors for fed-batch fermentations.
Fed-batch fermentations involve feeding sugar or other nutrients continuously or semi-continuously into a bioreactor.
As opposed to batch fermentation — where media is added to a bioreactor only once at the beginning of the experiment — fed-batch fermentation uses pumps to consistently supply the cells with fresh nutrients, allowing them to achieve higher productivity.
However, most fermentation processes require a gradual increase of nutrients across several hours, often overnight and during weekends, which isn’t practical to do manually.
“We were not able to adequately supply nutrients to our bioreactors between class sessions, which resulted in low yield in our fed-batch fermentation experiments,” Bruce said.
To address this, ABPDU developed a software application that allows labs similar to Laney’s teaching lab to control feed pumps conveniently and remotely.
“Users can load in spreadsheets that contain a few simple instructions for a programmable pump in the form of a timetable and pump speeds. The software then goes through the instructions line by line, executing whatever you tell the system to do,” said ABPDU senior process engineer Jan-Philip (JP) Prahl, who led the effort alongside ABPDU’s control expert Matt Stettler. “Users can also remotely access the computer in case something needs to be altered — a clear advantage when dealing with several day-long cultivations and limited resources.”
The software was made available to Laney College’s Biomanufacturing Program, and is now also available to the public.

“This pump will allow our students to design feeding schedules and even remotely control the feeding of our cells,” Bruce said. “In addition, we can now teach important concepts around the economic viability of bioprocesses.”
This capability brings Laney’s lab up to speed with industry standards.
“Fermentation is quickly becoming a standard piece of equipment in labs around the Bay Area and in innovative biotech in general,” said Ron Shigeta, a biotech mentor and co-founder of IndieBio.“ABPDU has helped to automate our fermentor stack with this project, setting the students to the current standards of industry practices.”